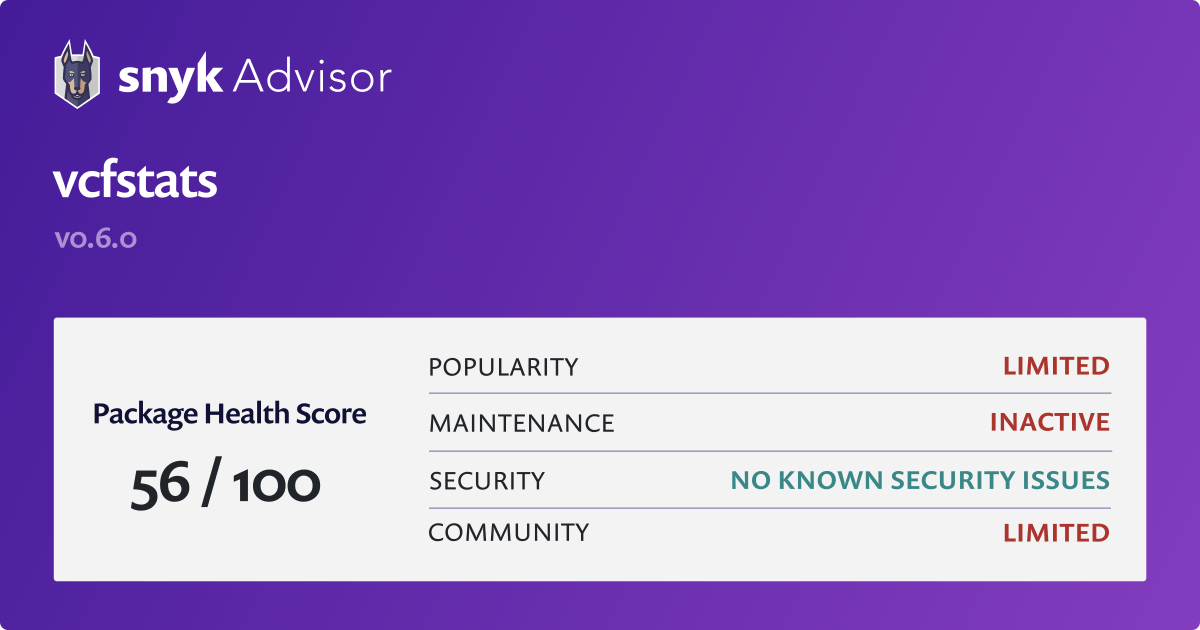
ubuntu - I tried many time to install requests python module but I cant, how I fix the path error on linux pop os? - Stack Overflow
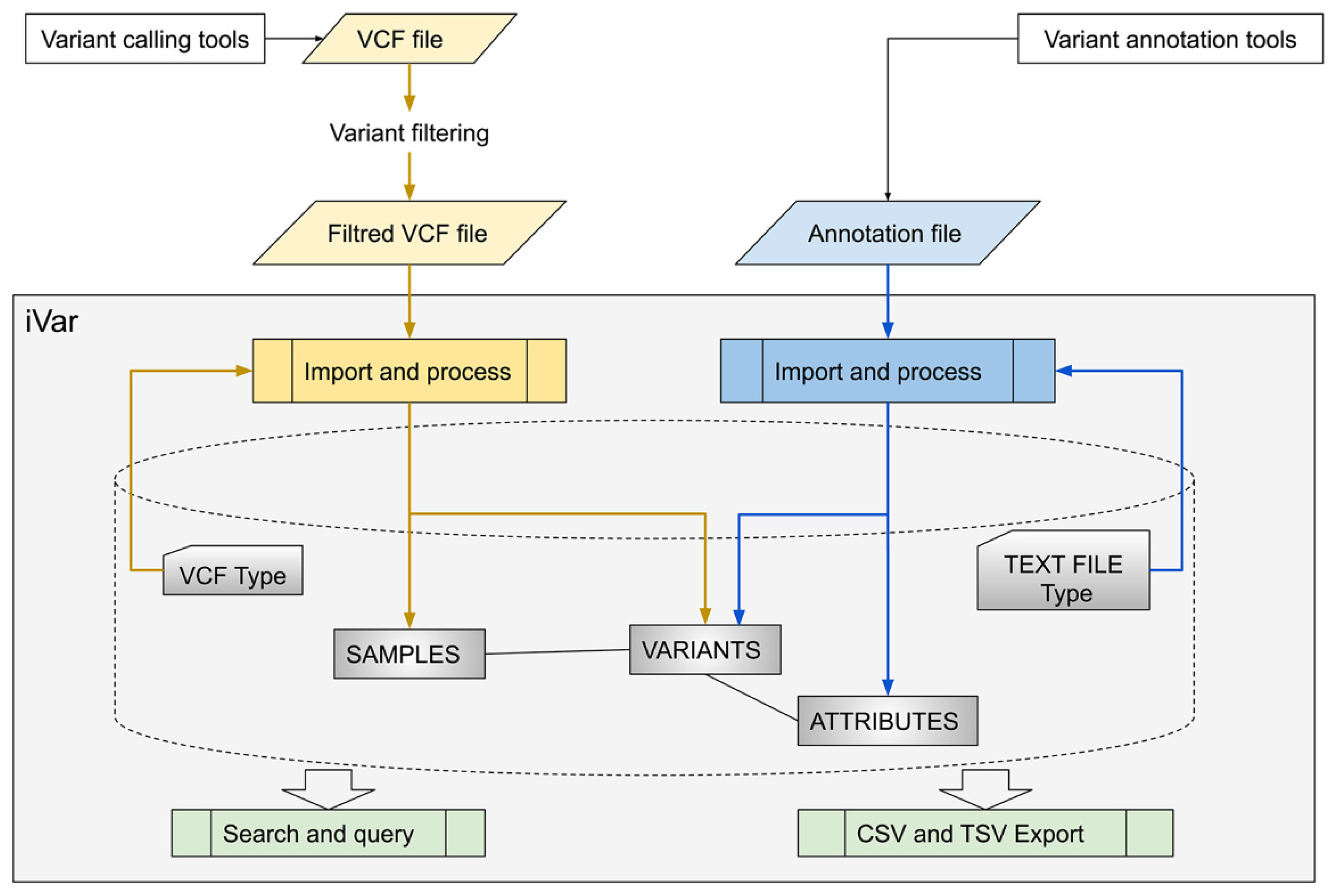
Genes | Free Full-Text | iVar, an Interpretation-Oriented Tool to Manage the Update and Revision of Variant Annotation and Classification

VCF‐Server: A web‐based visualization tool for high‐throughput variant data mining and management - Jiang - 2019 - Molecular Genetics & Genomic Medicine - Wiley Online Library
GitHub - iqbal-lab-org/cluster_vcf_records: Python package to cluster VCF records. Used by gramtools and minos

Linux for Bioinformatics | How to count the number variants in a VCF file | Beginners Course - YouTube
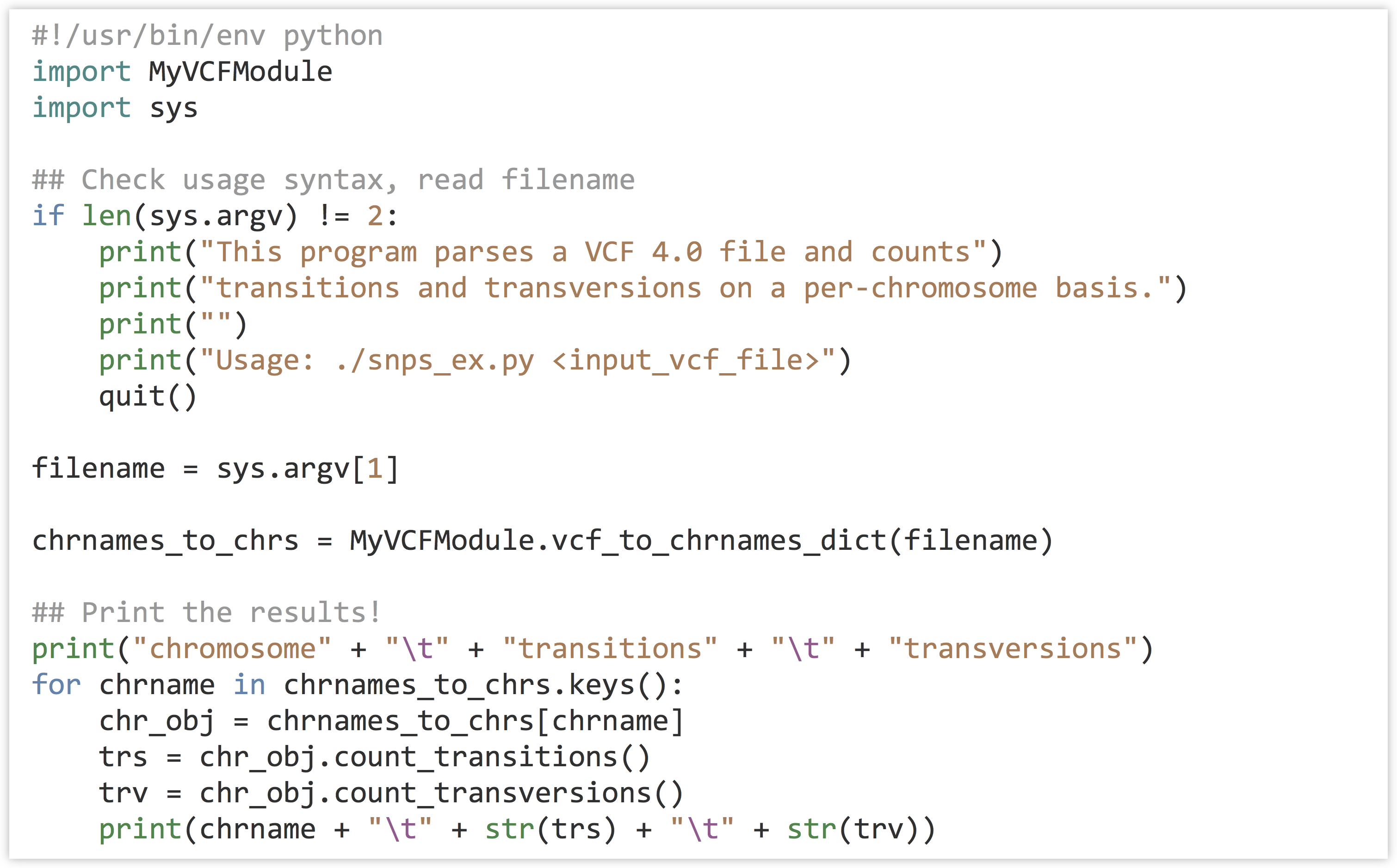
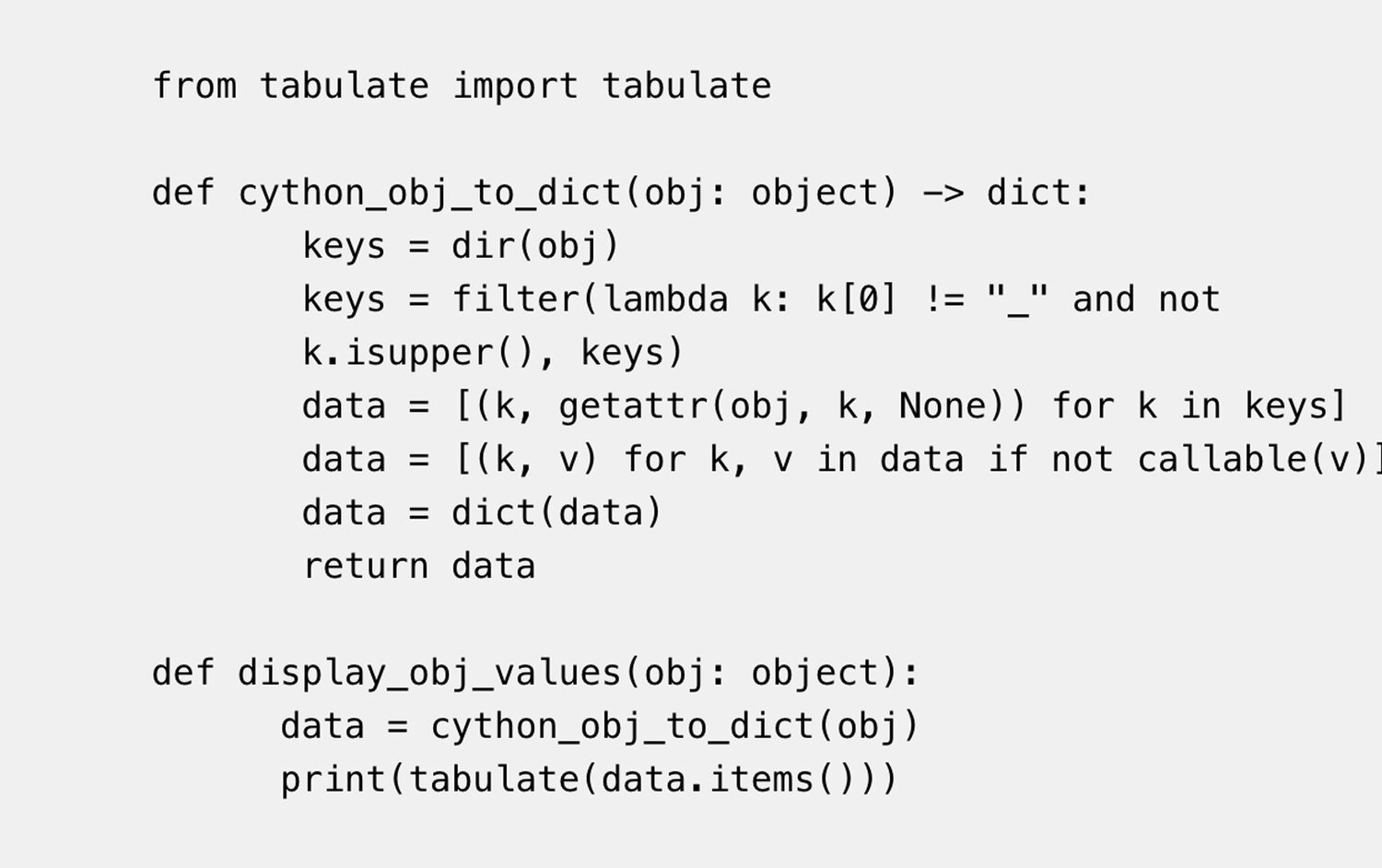
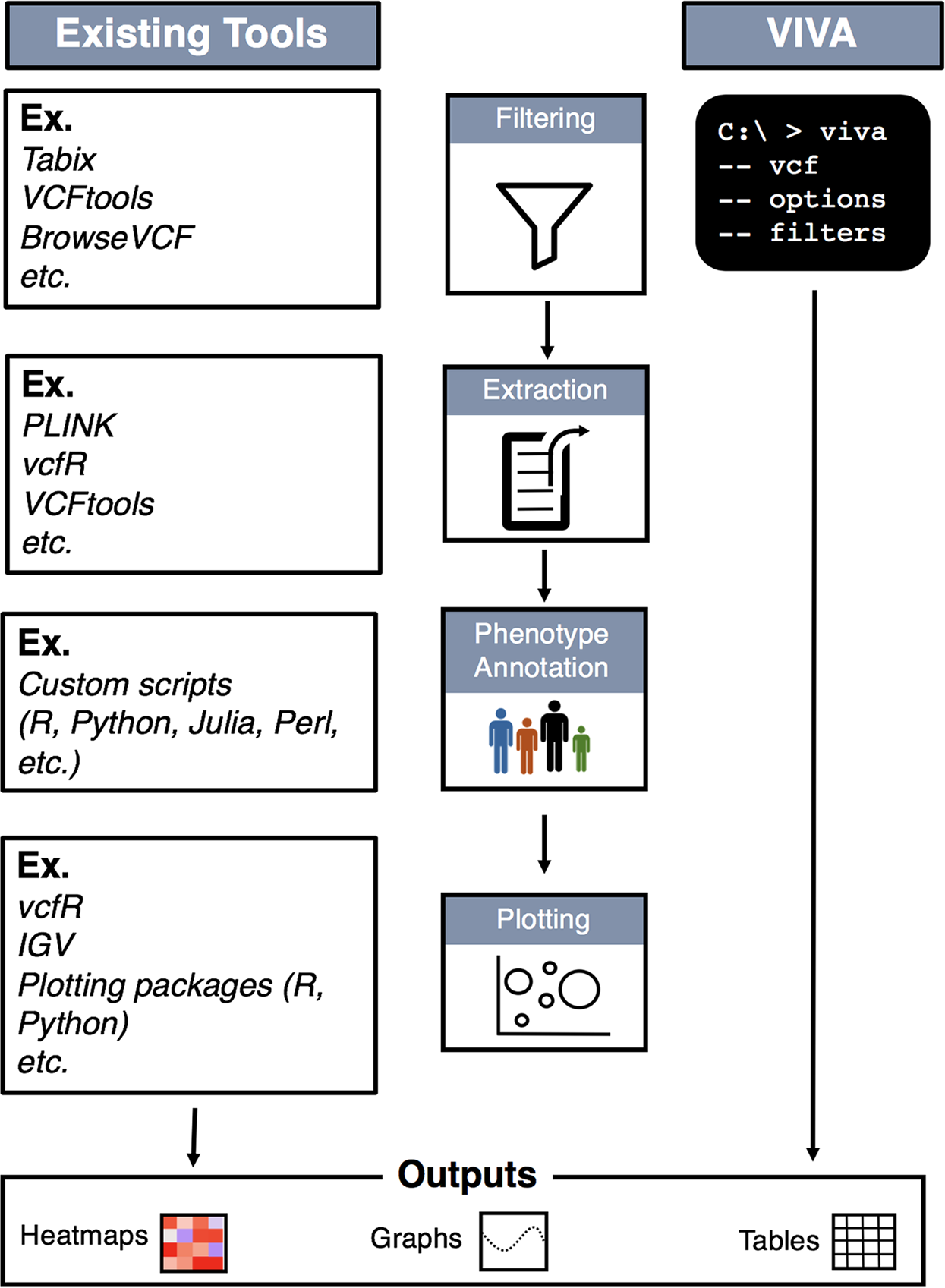
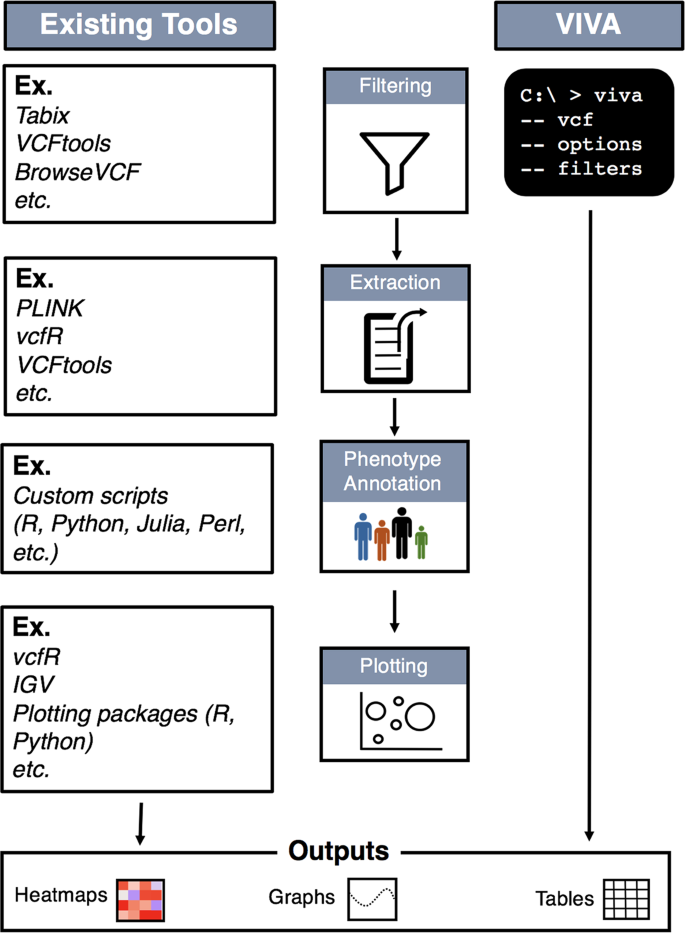
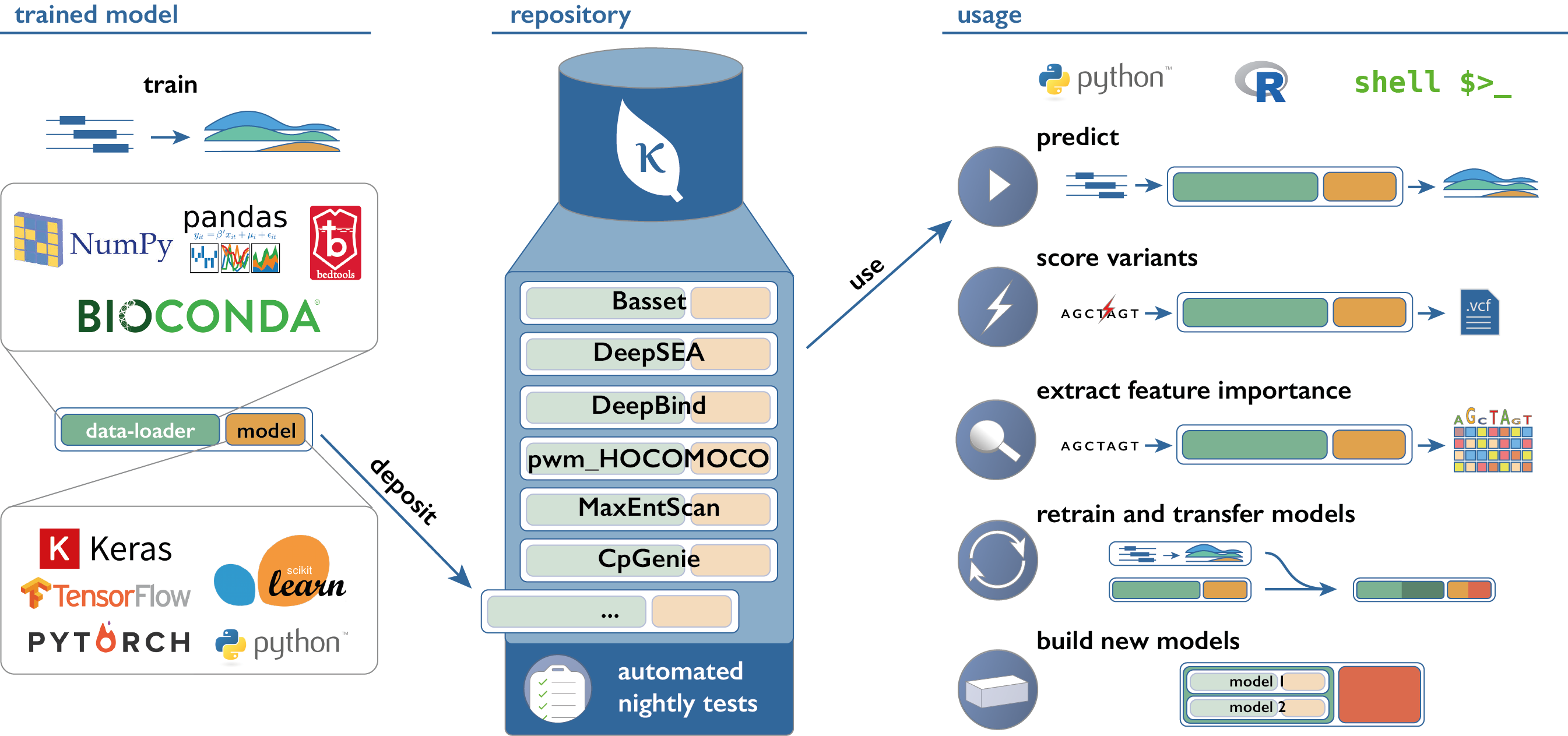
Application Programming Interfaces, Modules, Packages, Syntactic Sugar – A Primer for Computational Biology
![Python] VCF 파일을 데이터프레임(DataFrame)으로 Python] VCF 파일을 데이터프레임(DataFrame)으로](https://blog.kakaocdn.net/dn/bBCAS1/btqHLfHJboa/sb56yrSZ7zNyh7oEYMmju0/img.png)